Calcium imaging can record the neural activity of living animals in parallel with single-cell resolution, which provides the possibility to decipher the transmission, integration and computerization of information in neural circuits. In order to perform accurate neurological function analysis, it is particularly critical to obtain calcium imaging data with a high signal-to-noise ratio. However, due to the low peak accumulation and fast dynamic characteristics of calcium transient in vivo [1,2], the detector cannot capture enough fluorescent photons, and it has been difficult to break through the Poisson noise limit caused by the quantum nature of light. The problem of low signal-to-noise ratio in calcium imaging has long plagued neuroscientists. The existing methods have to sacrifice imaging speed, resolution or tissue activity in exchange for image signal-to-noise ratio, which greatly limits the development of life sciences.On August 16th, 2021, Nature Methods published online a research paper entitled Reinforcing neuron extraction and spike inference in calcium imaging using deep self-supervised denoising by Dai Qionghai's research group from the Institute of Brain and Cognitive Sciences and Department of Automation, Tsinghua University. This paper proposes a self-supervised learning method for calcium imaging denoising (DeepCAD), which uses only a single calcium imaging video sequence with a low signal-to-noise ratio to achieve network training.

Due to the characteristics of the high-speed and non-repetitiveness of neuronal calcium activity, the training truth value necessary for traditional supervised learning cannot be obtained. This self-supervised learning strategy breaks the dependence on training truth value and makes a breakthrough in calcium imaging denoising. On this basis, DeepCAD uses a three-dimensional network structure to make full use of the spatio-temporal correlation of neural activity to increase the signal-to-noise ratio of calcium imaging by more than ten times, and overcome the limit of photon noise. This method is used to observe the calcium dynamics of the neural fiber network and neuron community in the cerebral cortex of mouse, realizes calcium imaging with high signal-to-noise ratio under low excitation power, and is widely applicable to various calcium imaging systems.
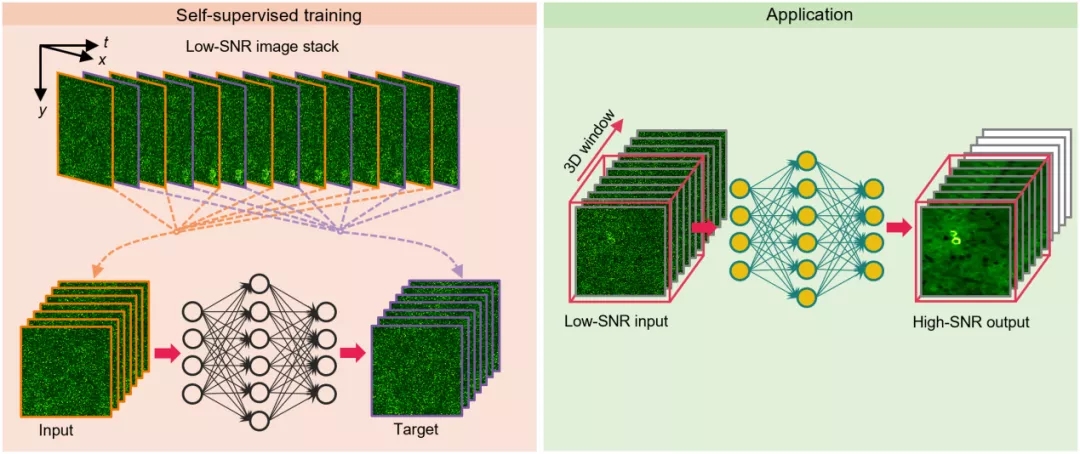
Figure 1. DeepCAD principle and application schematic diagram
In order to verify the accuracy and reliability of the method, the author built a special two-photon microscope. The fluorescent photons emitted by the sample are divided into two parts of 1:10 by the spectrometer at the detection end, and the two signals are collected simultaneously. The image with low signal-to-noise ratio is used as raw data input to the network, and the image with high signal-to-noise ratio is used to verify the output result. DeepCAD was first used for the calcium imaging denoising of Layer 1 neural fiber network in the mouse cerebral cortex. The results showed that the signal-to-noise ratio of the data after denoising was increased by more than ten times, and the calcium activity that was originally submerged in the noise could be truly restored.
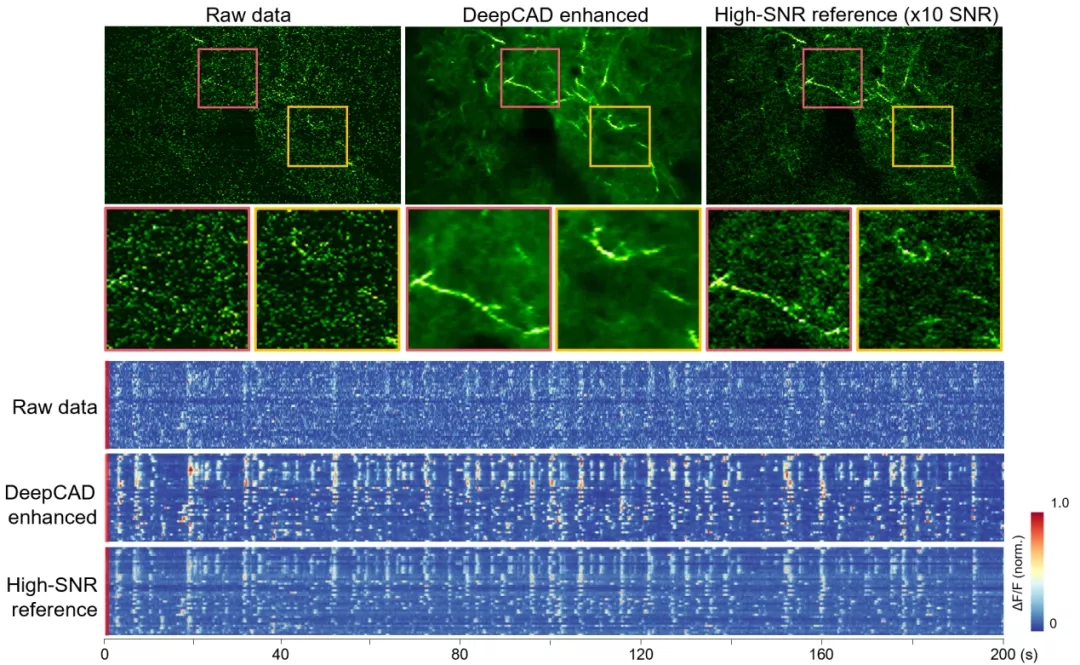
Figure 2. DeepCAD restores the calcium dynamics of the nerve fiber network submerged in noise, and the signal-to-noise ratio is increased by more than ten timesDeepCAD has also been used to observe the calcium activity of Layer 2/3 neural communities in the cerebral cortex of mice. The calcium imaging data after denoising has a very high signal-to-noise ratio, and the shape and distribution of neurons can be clearly identified from only a single frame of image. Because the detection noise is effectively removed, the accuracy of the calcium signal of the cell body is also greatly improved. Quantitative analysis shows that the calcium imaging data enhanced by DeepCAD can significantly improve the accuracy of neuron extraction and spike inference, which helps to fully reveal neural circuit activities and avoid information loss caused by noise.
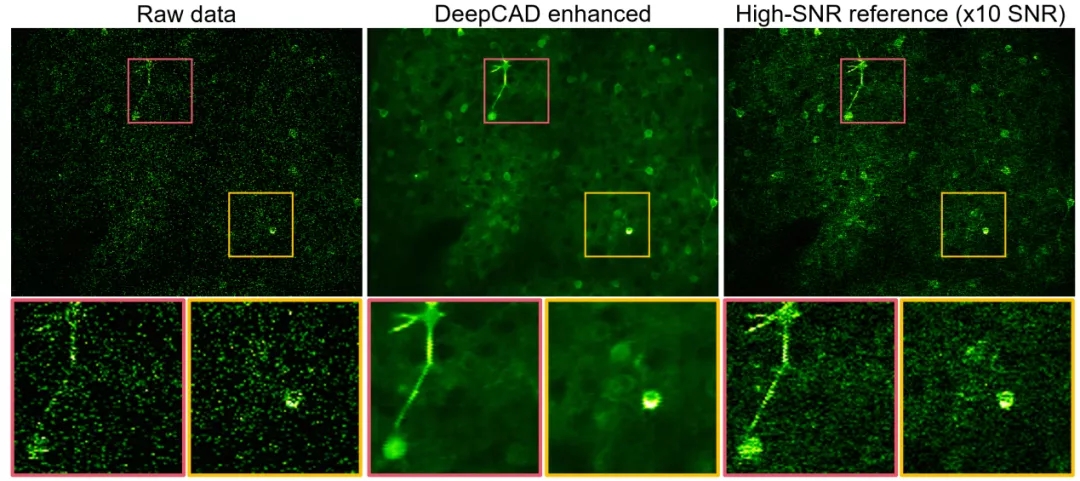
Figure 3. DeepCAD is used to denoise the calcium imaging of neural communities and accurately analyze neuronal activity
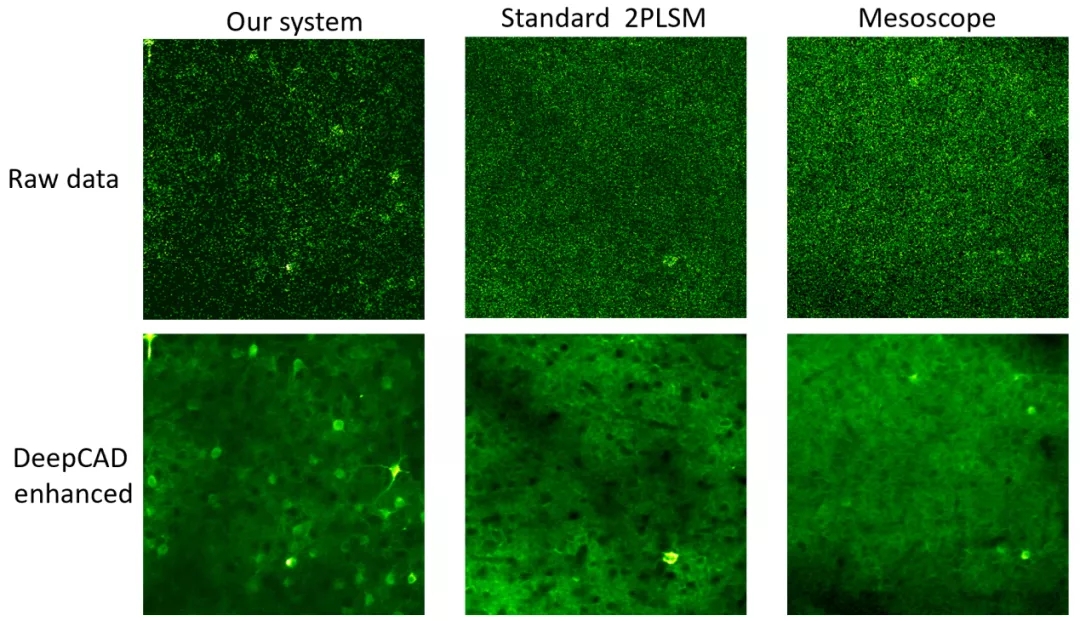
Finally, the author applies the pre-trained DeepCAD model to multiple two-photon microscopes equipped with different objective lenses and detectors. The results show that DeepCAD has good generalization and scalability, and it shows superior performance on different two-photon imaging systems.Figure 4. DeepCAD shows superior performance on different two-photon microscopesIn order to simplify the use of DeepCAD, the author encapsulated the method as a Fiji plug-in. At present, relevant codes, plug-ins and complete data sets have been released along with the paper. In addition to nerve calcium imaging, this method is expected to be extended to other imaging modes, such as wide-field microscope, confocal microscope, and light sheet microscope, or other functional imaging scenarios, such as cell migration observation and voltage imaging. DeepCAD can be used as a general data processing step for time-lapse imaging in the case of photon limitation to achieve low excitation power and long-term observation of biological dynamic activities.
Figure 5. Fiji plugin for DeepCADDoctoral students Li Xinyang and Zhang Guoxun from the Department of Automation of Tsinghua University are the co-first authors of the paper; Professor Dai Qionghai from the Institute of Brain and Cognitive Sciences of Tsinghua University and the Department of Automation of Tsinghua University, Professor Wang Haoqian from the Shenzhen International Graduate School of Tsinghua University and Associate Professor Fang Lu from the Department of Electronics of Tsinghua University are the co-corresponding authors of the paper.Original link:
https://www.nature.com/articles/s41592-021-01225-0
Paper code link:
https://github.com/cabooster/DeepCAD
1 Sabatini, B. L., Oertner, T. G. & Svoboda, K. The Life Cycle of Ca2+ Ions in Dendritic Spines. Neuron 33, 439-452, doi:10.1016/s0896-6273(02)00573-1 (2002).2 Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295-300, doi:10.1038/nature12354 (2013).